From Structure to Solution: How MacMolecule 2 Visualized HIV Protease and Its Inhibitors

What Is HIV?
HIV (Human Immunodeficiency Virus) is a retrovirus that attacks the body’s immune system, specifically the CD4+ T lymphocytes (T helper cells). These cells are crucial for mounting effective immune responses against infections and diseases.
If left untreated, HIV gradually destroys these cells, weakening the immune system and making the body vulnerable to opportunistic infections and certain cancers. Advanced HIV infection can lead to AIDS (Acquired Immunodeficiency Syndrome), which is the final stage of HIV disease.
Structure
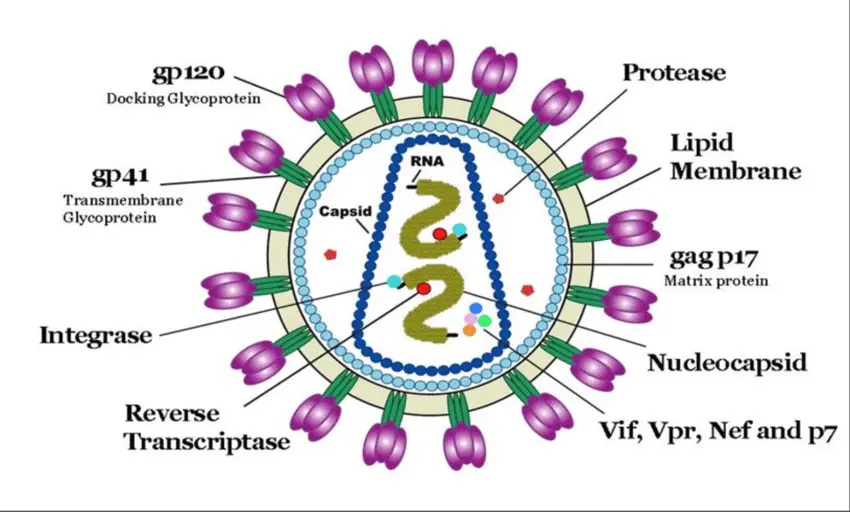
It contains two identical single-stranded RNA genomes unusual because most organisms use DNA as their genetic material.
Inside the envelope are viral proteins, including reverse transcriptase, integrase, and HIV protease key enzymes that allow the virus to replicate.
HIV is an enveloped virus with an outer lipid membrane derived from the host cell.
Genome
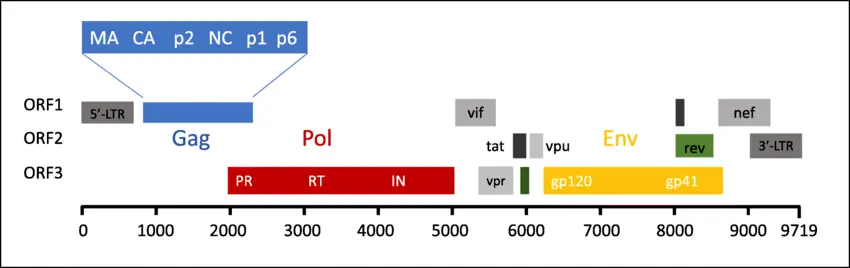
The HIV genome is small (~9.7 kb) but highly efficient.
It contains genes like:
- gag encodes structural proteins.
- pol encodes enzymes: reverse transcriptase, integrase, protease.
- env encodes envelope glycoproteins (gp120 and gp41) for binding and fusion with host cells.
- regulatory genes e.g., tat, rev that modulate viral replication.
Life Cycle
The HIV replication cycle involves several unique steps:
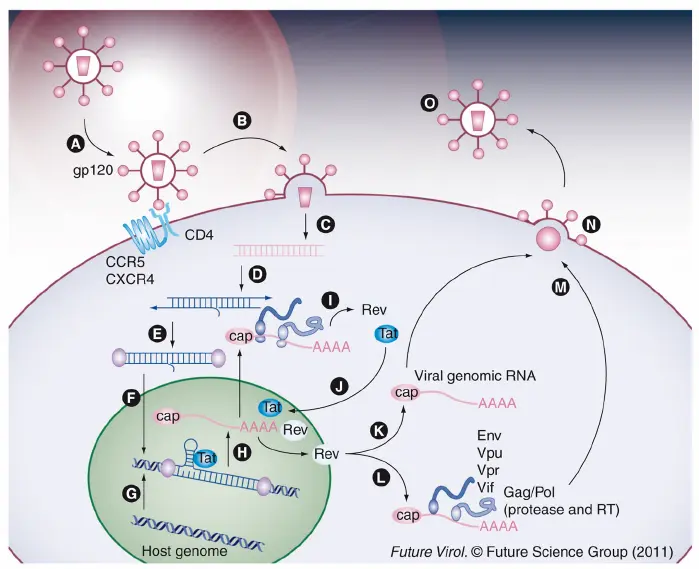
- Attachment and Entry: The virus binds to the CD4 receptor and co-receptors (CCR5 or CXCR4) on the host cell.
- Fusion: The viral envelope fuses with the host cell membrane, releasing viral RNA and enzymes.
- Reverse Transcription: HIV’s reverse transcriptase converts viral RNA into DNA this is a key step that distinguishes retroviruses.
- Integration: The viral DNA enters the host nucleus and is integrated into the host genome by the integrase enzyme.
- Transcription and Translation: Host cell machinery transcribes and translates viral genes, producing viral proteins.
- Assembly and Maturation: New virus particles assemble at the cell membrane. HIV protease cleaves polyproteins to form mature, infectious virions.
- Budding: New viruses bud off to infect other cells.

Role in the HIV Life Cycle
- After HIV infects a host cell, its genetic material is reverse-transcribed into DNA and integrated into the host genome.
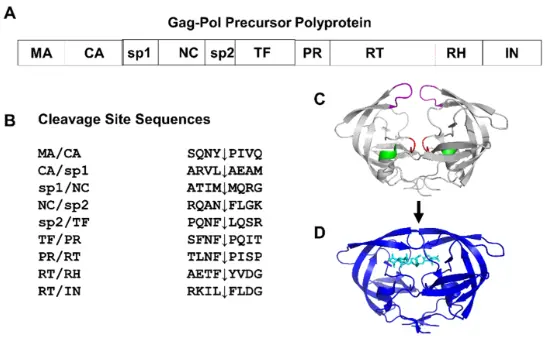
- The viral DNA directs the host cell to produce large polyprotein precursors.
- HIV protease cleaves these long polyproteins into functional smaller proteins structural proteins and enzymes needed to assemble mature, infectious viral particles.
- Without an active protease, HIV cannot mature and remains non-infectious.
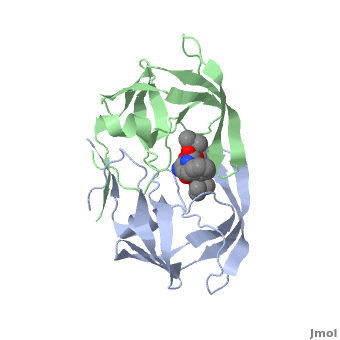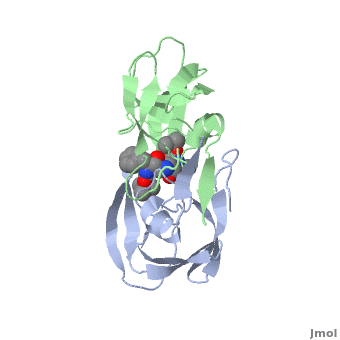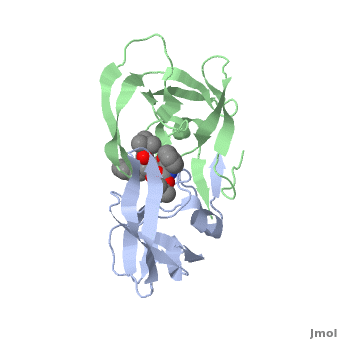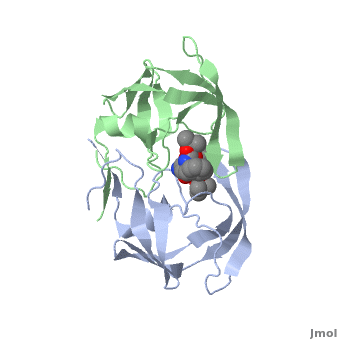
Why It’s a Therapeutic Target
Because of its essential role, HIV protease became one of the first viral enzymes targeted for antiretroviral therapy. Protease inhibitors (PIs) bind to the active site of the enzyme, blocking its ability to process viral proteins, thus halting viral replication.
Structure–Function Relationships
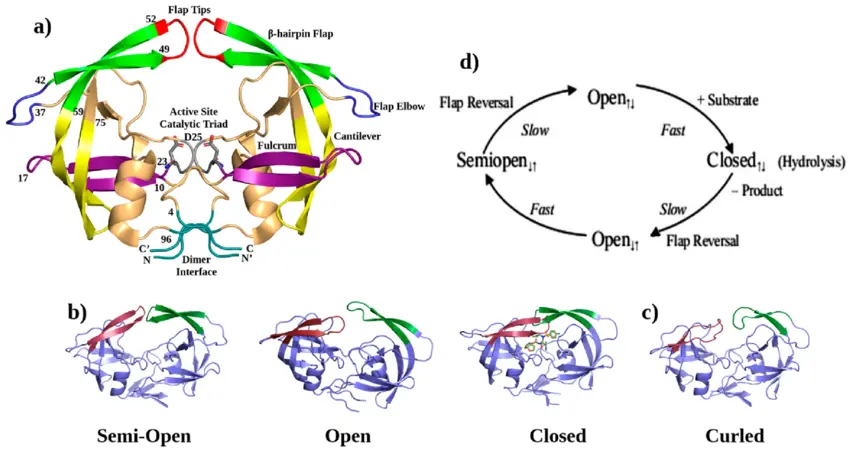
The structure of HIV protease was one of the earliest viral enzyme structures solved by X-ray crystallography in the late 1980s. This revealed:
- It is a homodimer two identical subunits forming a symmetrical enzyme.
- The active site sits between the subunits.
- A flexible “flap” region covers the active site, opening and closing to allow substrate or inhibitor binding.
This structural insight directly enabled structure-based design of the first generation of protease inhibitors such as saquinavir, indinavir, and ritonavir, all of which significantly improved outcomes for individuals living with HIV.

Bridging Data and Discovery: The Role of Visualization Tools
Molecular visualization tools allow researchers and students to explore molecular structures in three dimensions. Instead of abstract letters on a page, they can see how helices, sheets, and active sites are arranged in space.
This is where tools like MacMolecule 2 and PCMolecule 2 come into the story.

In the era when these structures were first published, having access to detailed 3D molecular data was a scientific revolution but researchers needed tools to visualize and analyze this data.
MacMolecule 2 (for Macintosh) and PCMolecule 2 (or similar PC-based versions) were among the pioneering desktop molecular graphics applications in the 1990s. They allowed researchers, educators, and students to:
Open PDB files (Protein Data Bank files) of HIV protease and its inhibitor complexes.
View the active site in three dimensions, highlighting key amino acids and how they interact with target molecules.
Rotate and zoom the structure to study the flap region, binding pockets, and hydrogen bonds.
Generate images for publications and presentations, making structural biology findings more accessible.
Impact on Research, Education, and Therapeutic Development
By enabling researchers to explore HIV protease–inhibitor complexes on personal computers, these early visualization tools democratized access to structural biology. They:
Supported rational design, enabling chemists to modify inhibitor structures for a more precise fit within the active site.
Helped explain resistance mechanisms, as mutations in the protease alter its structure and affect how inhibitors bind.
Enhanced teaching and training, enabling students to see real examples of how protein structure influences the action of therapeutic inhibitors.
The Legacy and Modern Advances